Statistics¶
Collection of tools for calculating statistics.
CDF¶
Cumulative Distribution Function¶
-
pygeostat.statistics.cdf.cdf(var, wt=None, lower=None, upper=None, bins=None)¶ Calculates an empirical CDF using the standard method of the midpoint of a histogram bin. Assumes that the data is already trimmed or that iwt=0 for variables which are not valid.
If ‘bins’ are provided, then a binned histogram approach is used. Otherwise, the CDF is constructed using all points (as done in GSLIB).
Notes
‘lower’ and ‘upper’ limits for the CDF may be supplied and will be returned appropriately
Parameters: - var (array) – Array passed to the cdf function
- wt (str) – column where the weights are stored
- Lower (float) – Lower limit
- upper (float) – Upper Limit
- bins (int) – Number of bins to use
Returns: - midpoints (np.ndarray) – array of bin midpoints
- cdf (np.ndarray) – cdf values for each midpoint value
wtpercentile¶
-
pygeostat.statistics.cdf.wtpercentile(cdf_x, cdf, percentile)¶ Given ‘x’ values of a CDF and corresponding CDF values, find a given percentile. Percentile may be a single value or an array-like and must be in [0, 100] or CDF bounds
z_percentile¶
-
pygeostat.statistics.cdf.z_percentile(z, cdf_x, cdf)¶ Given ‘cdf_x` values of a CDF and corresponding
cdfvalues, find the percetile of a given valuez. Percentile may be a single value or an array-like and must be in [0, 100] or CDF bounds.
Discretize CDF¶
-
pygeostat.statistics.cdf.discretize_cdf(nthresholds, data, wt=None, lower=None, upper=None)¶ Function to discretize a the cdf of the given dataset by equal probability intervals producing the midpoint x and y locations and the threshold value for each of the bins.
Notes
Functionality for choosing the thresholds should be added in the future, eg. a parameter thresholds containing a list of thresholds and then return the coordinates of the midpoints and y-values for the thresholds
Parameters: - nthresholds (int) – number of thresholds to generate, equally spaced in probability
- data (np.ndarray) – an array of data from which to build the cdf
- wt (np.ndarray) – weights to consider for building the cdf
- lower (float) – lower trimming value
- upper (float) – upper trimming value
Returns: mp_x, mp_y, zc_x, zc_y – lists of x and y coordinates for the midpoints (mp) and threshold (zc) for each bin
Return type: list
Examples
Discretize the cdf with 16 thresholds:
>>> (mp_x, mp_y), (cutoff_x, cutoff_y) = gs.discretize_cdf(16, df.data['Zn'])
Plot the cdf using
gs.histplt:>>> ax = gs.histplt(df.data['Zn'], icdf=True, figsize=(5, 4), xlim=(0, 60))
Plot the cutoff points on the cdf:
>>> ax.scatter(cutoff_x, cutoff_y, marker='+', s=10, lw=0.5, c='red')
Add some lines to show where the bins fall on the x-axis:
>>> for cutoff, yval in zip(cutoff_x, cutoff_y): ... ax.vlines(cutoff, 0, yval, colors='r', lw=0.25) ... ax.text(cutoff-0.1, yval + 0.001, round(cutoff,2), va='bottom', ha='right', ... fontsize=8)
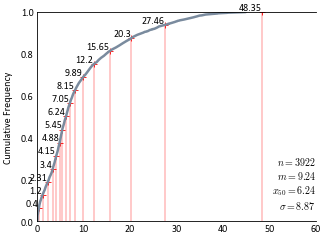
Code author: Ryan Martin - 2017-03-26
Smooth CDF¶
-
pygeostat.statistics.cdf.smooth_cdf(global_data, weights=None, ik_cdf_x=None, ik_cdf_y=None, lower=None, upper=None, nbins=500, windowfrac=None)¶ This function smooths the cdf of the given dataset using convolution with the given windowfrac parameter. Optionally a local cdf can be passed, and the smoothed distribution will be interpolated between the given cutoffs obtained from the ik3d distribution.
Parameters: - global_data (np.ndarray) – number of thresholds to generate, equally spaced in probability
- weights (np.ndarray) – weights for the global distribution, e.g. declustering weights
- ik_cdf_x (np.ndarray, optional) – cdf x-values corresponding to the output from
cdffor a distribution to smooth the global data too, e.g. interpolate the global to the MIK cutoffs - ik_cdf_y (np.ndarray, optional) – cdf y-values corresponding to the output from
cdffor a distribution to smooth the global data too, e.g. interpolate the global to the MIK cutoffs - lower (float) – the lower trimming limit for global_data; values below are omitted
- upper (float) – the upper trimming limit for global_data; values above are omitted
- nbins (int) – the number of interpolated values to return
- windowfrac (float) – the fraction of the total number of bins used in the window, e.g. 50% for nbins = 100 means that the window size is 50
- function (string) – permissable scipy.interpolate.Rbf functions, e.g.
'linear','cubic','multiquadric'
Returns: x, y – lists of x, y coordinates defining the CDF for the given nbins
Return type: list
Examples
Make some noisy data and smooth the CDF
>>> dataset = np.random.randn(100) + np.random.rand(100) * 0.5 >>> ax = gs.histplt(dataset, icdf=True) >>> colors = gs.get_palette('cat_pastel', 6).colors[2:] >>> for i, window in enumerate([5, 10, 25, 50]): ... smoothx, smoothy = gs.smooth_cdf(dataset, windowfrac=window) ... ax.plot(smoothx, smoothy, c=colors[i], lw=0.5, label='window=%i' % window) >>> ax.legend(loc='upper left', prop={'size':5})
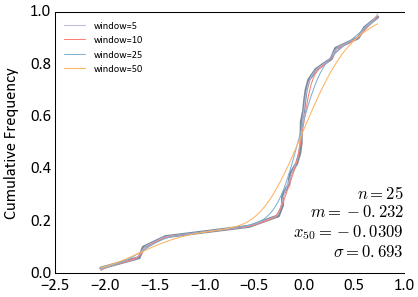
Try again with an example of MIK intervals and interpolating a CDF to the thresholds
>>> ax = gs.histplt(dataset, icdf=True, label='global cdf', figsize=(5,4), lower=0, ... bins=100) >>> probs = np.array([0, 0.45, 0.65, 0.67, 0.77, 1.0]) >>> zvals = np.array([0, 5, 15, 18, 22, 35]) >>> itpx, itpy = gs.smooth_cdf(dataset, ik_cdf_x=zvals, ik_cdf_y=probs, nbins=100, lower=0, ... windowfrac=0) >>> smoothx, smoothy = gs.smooth_cdf(dataset, ik_cdf_x=zvals, ik_cdf_y=probs, nbins=100, ... lower=0, windowfrac=7) >>> # local indicator CDF >>> pts, ipts = gs.build_indicator_cdf(probs, zvals) >>> ax.scatter(ipts[:, 0], ipts[:, 1], c='red', marker='o', label='cutoff midpoints') >>> ax.scatter(pts[:, 0], pts[:, 1], c='black', marker='+', label='cat cutoffs', zorder=5) >>> ax.plot(smoothx, smoothy, c=colors[0], label='smoothed cdf') >>> ax.plot(itpx, itpy, c=colors[3], label='rough cdf') >>> ax.plot(pts[:, 0], pts[:, 1], c=colors[2], zorder=0, label='cat cdf') >>> ax.set_xbound(lower=0) >>> ax.set_ybound(lower=0, upper=1) >>> ax.legend(loc='center right', prop={'size': 6})

Code author: Ryan Martin - 2017-04-17
Build Indicator CDF¶
-
pygeostat.statistics.cdf.build_indicator_cdf(prob_ik, zvals)¶ Build the X-Y data required to plot a categorical cdf
Parameters: - prob_ik (np.ndarray) – the p-vals corresponding to the cutoffs
- zvals (np.ndarray) – the corresponding z-value specifying the cutoffs
Returns: points, midpoints – the x and y coordinates of (1) the cutoffs and (2) the midpoints for each cutoff
Return type: np.ndarray
Code author: Ryan Martin - 2017-04-17
Kernel Density Estimation Functions¶
Univariate KDE with StatsModels¶
-
pygeostat.statistics.kde.kde_statsmodels_u(x, x_grid, bandwidth=0.2, **kwargs)¶ Univariate Kernel Density Estimation with Statsmodels
Multivariate KDE with StatsModels¶
-
pygeostat.statistics.kde.kde_statsmodels_m(x, x_grid, bandwidth=0.2, **kwargs)¶ Multivariate Kernel Density Estimation with Statsmodels
KDE with Scikit-learn¶
-
pygeostat.statistics.kde.kde_sklearn(x, x_grid, bandwidth=0.2, **kwargs)¶ Kernel Density Estimation with Scikit-learn
KDE with Scipy¶
-
pygeostat.statistics.kde.kde_scipy(x, x_grid, bandwidth=0.2, **kwargs)¶ Kernel Density Estimation based on different packages and different kernels Note that scipy weights its bandwidth by the covariance of the input data. To make the results comparable to the other methods, we divide the bandwidth by the sample standard deviation here.
Weighted Statistics¶
Weighted Correlation¶
-
pygeostat.statistics.utils.weightedcorr(x, y, wt)¶ Calculates the weighted correlation
Weighted Rank Correlation¶
-
pygeostat.statistics.utils.weightedcorr_rank(x, y, wt)¶ Calculatees the weighted spearman rank correlation
Assorted Stats Functions¶
Nearest Positive Definite Correlation Matrix¶
-
pygeostat.statistics.utils.nearpd(inmat)¶ This function uses R to calculate the nearest positive definite matrix within python. An installation of R with the library “Matrix” is required. The module rpy2 is also needed
The only requirement is an input matrix. Can be either a pandas dataframe or numpy-array.
Parameters: inmat (np.ndarray, pd.DataFrame) – input numpy array or pandas dataframe, not numpy matrix Returns: pdmat – Nearest positive definite matrix as a numpy-array Return type: np.ndarray Code author: Warren Black - 2015-08-02
Accuracy Plot Statistics - Simulation¶
-
pygeostat.statistics.utils.accsim(truth, reals, pinc=0.05)¶ Calculates the proportion of locations where the true value falls within symmetric p-PI intervals when completing a jackknife study. A portion of the data is excluded from the conditioning dataset and the excluded sample locations simulated values are then checked.
See also
Pyrcz, M. J., & Deutsch, C. V. (2014). Geostatistical Reservoir Modeling (2nd ed.). New York, NY: Oxford University Press, p. 350-351.
Parameters: - truth (np.ndarray) – Tidy (long-form) 1D data where a single column containing the true values. A pandas dataframe/series or numpy array can be passed
- reals (np.ndarray) – Tidy (long-form) 2D data where a single column contains values from a single realizations and each row contains the simulated values from a single truth location. A pandas dataframe or numpy matrix can be passed
- pinc (float) – Increments between the probability intervals to calculate within (0, 1)
Returns: - propavg (pd.DataFrame) – Dataframe with the calculated probability intervals and the fraction within the interval
- sumstats (dict) – Dictionary containing the average variance (U), mean squared error (MSE), saccuracy measure (acc), precision measure (pre), and a goodness measure (goo)
Code author: Warren Black - 2016-07-21
Accuracy Plot Statistics - CDF thresholds¶
-
pygeostat.statistics.utils.accmik(truth, thresholds, mikprobs, pinc=0.05)¶ Similar to accsim but accepting mik distributions instead
Mostly pulled from accsim
Parameters: - truth (np.ndarray) – Tidy (long-form) 1D data where a single column containing the true values. A pandas dataframe/series or numpy array can be passed
- thresholds (np.ndarray) – 1D array of thresholds where each CDF is defined by these thresholds and the probability given in the mikprobs array for each location.
- mikprobs (np.ndarray) – Tidy (long-form) 2D data where a single column contains values from a single MIK cutoff and each row contains the simulated values for the corresponding single truth location. A pandas dataframe or numpy matrix can be passed
- pinc (float) – Increments between the probability intervals to calculate within (0, 1)
Returns: - propavg (pd.DataFrame) – Dataframe with the calculated probability intervals and the fraction within the interval
- sumstats (dict) – Dictionary containing the average variance (U), mean squared error (MSE), accuracy measure (acc), precision measure (pre), and a goodness measure (goo)
Code author: Warren Black and Ryan Martin - 2017-04-17
PostSim¶
-
pygeostat.statistics.postsim.postsim_multfiles(file_base_or_list, output_name, Nr=None, file_ending=None, fltype=None, output_fltype=None, zero_padding=0, variables=None, var_min=None)¶ The multiple file postsim function uses recursive statistics for memory management and coolness factor. See http://people.revoledu.com/kardi/tutorial/RecursiveStatistic/ This function will take multiple realizations and post process the results into mean and variance for each variable. You can either pass it a list of files to iterate through or a filebase name and the number of realizations.
Parameters: - file_base_or_list (list) or (str) – List of files or path + base name of sequentially named files
- output_name (str) – ath (or name) of file to write output to.
- Nr (int) – Number of realizations. Needed if file base name is passed.
- file_ending (str) – file ending (ex. “out”). Used if file base name is passed. Period is not included.
- fltype (str) – Type of data file: either
csv,gslib,hdf5, orgsb. Used if file base name is passed and file_ending is not used. - output_fltype (str) – Type of output data file: either
csv,gslib,hdf5, orgsb. - zero_padding (int) – Number of zeros to padd number in sequentially named files with. Default is 0.
- variables (str) – List of variables to process.
- var_min (list) or (float) – Minimum trimming limit to use. If one value is passed it will apply the trimming limit to all variables. Or a list of trimming limit for each variable can be passed.
Code author: Tyler Acorn - 2016-08-03